Clustering k-Means
2 Jan 2018
Introduction to k-Means
k-Means Algorithm allow to partition a set of training data in k clusters in which each observation belongs to the the cluster with the nearest mean. The result is a partition of table called Voronoi cells.
Algorithm to find clusters
This algoritm is called Lloyd’s algorithm:
Input:
- $K$ (number of clusters)
- Training set $\lbrace x^{(1)}, x^{(2)},… x^{(m)} \rbrace; z_i \in \mathbb{R}^n$
Randomy initialize K cluster centroids $\mu_1, \mu_2,…\mu_K \in \mathbb{R}^n$
Repeat {
for $i=1$ to $m$:
$c^{(i)}$ = index ( from 1 to K ) of cluster centroid closest to $x_i$
for $k=1$ to $K$:
$\mu_k$ = average (mean) of points assigned to cluster $k$
} until $c^{(i)}$ are the same as the previous iteration
Proces minimizes with this cost:
with respect to $C_k$, $\mu_k$
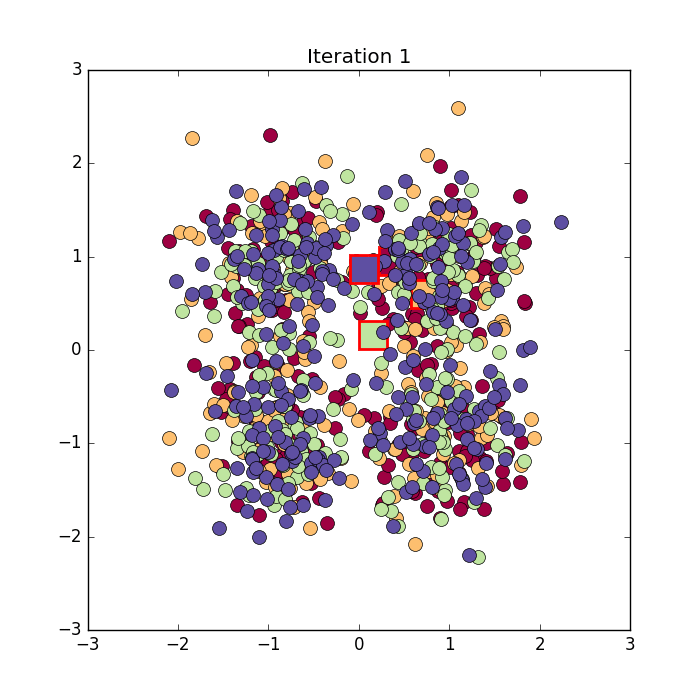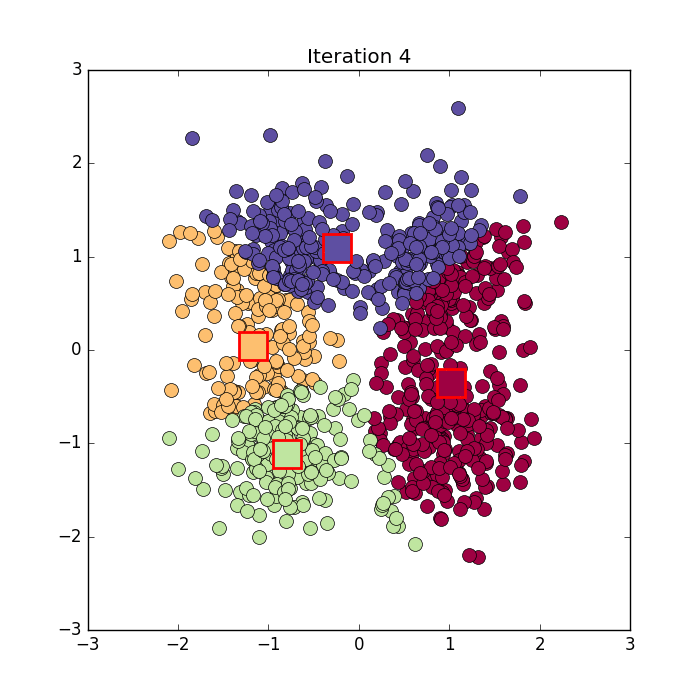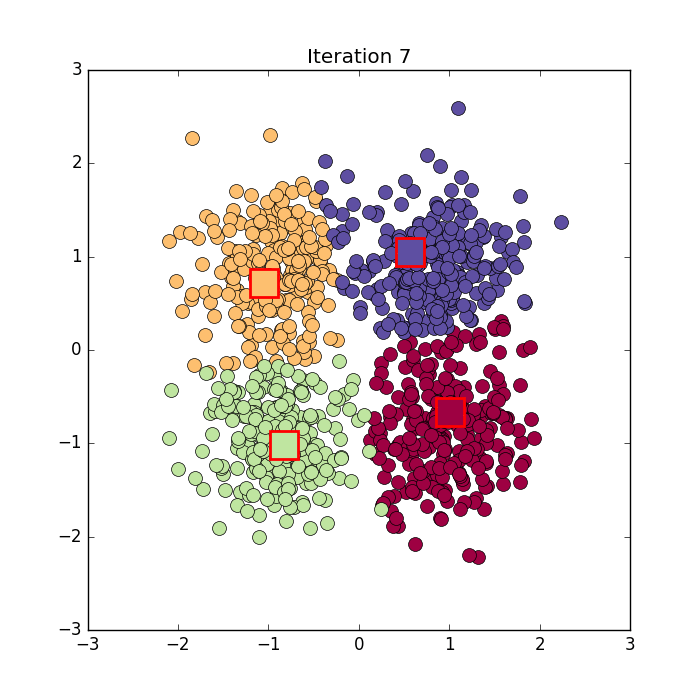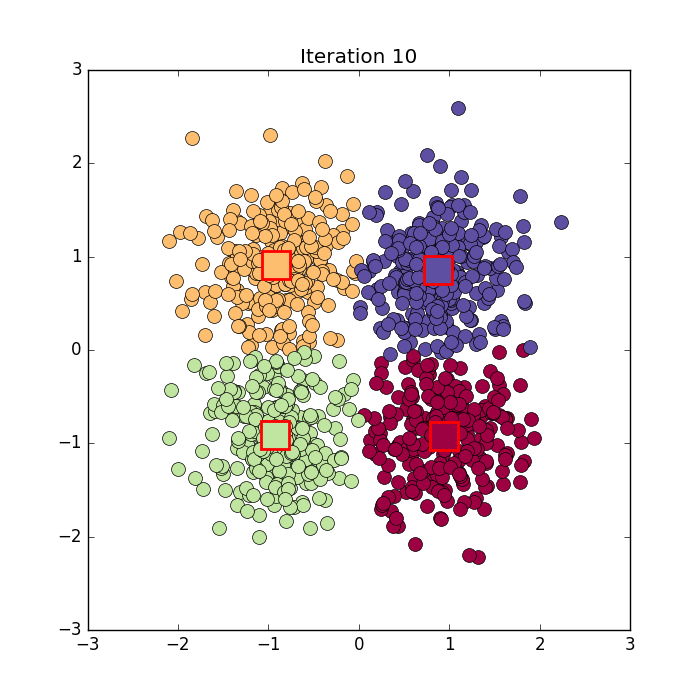
Algorithm in python from here:
import numpy as np
def cluster_points(X, mu):
clusters = {}
for x in X:
bestmukey = min([(i[0], np.linalg.norm(x-mu[i[0]])) \
for i in enumerate(mu)], key=lambda t:t[1])[0]
try:
clusters[bestmukey].append(x)
except KeyError:
clusters[bestmukey] = [x]
return clusters
def reevaluate_centers(mu, clusters):
newmu = []
keys = sorted(clusters.keys())
for k in keys:
newmu.append(np.mean(clusters[k], axis = 0))
return newmu
def has_converged(mu, oldmu):
return (set([tuple(a) for a in mu]) == set([tuple(a) for a in oldmu])
def find_centers(X, K):
# Initialize to K random centers
oldmu = random.sample(X, K)
mu = random.sample(X, K)
while not has_converged(mu, oldmu):
oldmu = mu
# Assign all points in X to clusters
clusters = cluster_points(X, mu)
# Reevaluate centers
mu = reevaluate_centers(oldmu, clusters)
return(mu, clusters)
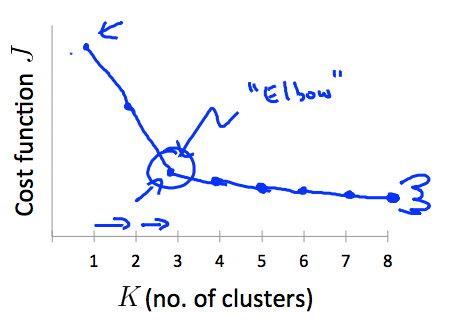
Choosing $K$
Elbow method:
“elbow” cannot always be unambiguously identified.
Gap statistic (extracted from here)
It’s a way to standarize to comparison. We evaluate the Gap from a random set of data from the k-Mean for every K.
$W^*_{kb}$ has been generated from a average of $B$ copies generated with Monte Carlo sample distribution.
standar deviation $sd(k)$:
Choose smallest $K$ that satisfies: $Gap(k) \ge Gap(k+1) - s_{k+1}$
We assume that we have computed the k-Mean result from the above algorithm:
def Wk(mu, clusters):
K = len(mu)
return sum([np.linalg.norm(mu[i]-c)**2/(2*len(c)) \
for i in range(K) for c in clusters[i]])
With these cluster results we find the K best aproach.
def find_centers(X, K):
# Initialize to K random centers
oldmu = random.sample(X, K)
mu = random.sample(X, K)
while not has_converged(mu, oldmu):
oldmu = mu
# Assign all points in X to clusters
clusters = cluster_points(X, mu)
# Reevaluate centers
mu = reevaluate_centers(oldmu, clusters)
return(mu, clusters)
def bounding_box(X):
xmin, xmax = min(X,key=lambda a:a[0])[0], max(X,key=lambda a:a[0])[0]
ymin, ymax = min(X,key=lambda a:a[1])[1], max(X,key=lambda a:a[1])[1]
return (xmin,xmax), (ymin,ymax)
def gap_statistic(X):
(xmin,xmax), (ymin,ymax) = bounding_box(X)
# Dispersion for real distribution
ks = range(1,10)
Wks = zeros(len(ks))
Wkbs = zeros(len(ks))
sk = zeros(len(ks))
for indk, k in enumerate(ks):
mu, clusters = find_centers(X,k)
Wks[indk] = np.log(Wk(mu, clusters))
# Create B reference datasets
B = 10
BWkbs = zeros(B)
for i in range(B):
Xb = []
for n in range(len(X)):
Xb.append([random.uniform(xmin,xmax),
random.uniform(ymin,ymax)])
Xb = np.array(Xb)
mu, clusters = find_centers(Xb,k)
BWkbs[i] = np.log(Wk(mu, clusters))
Wkbs[indk] = sum(BWkbs)/B
sk[indk] = np.sqrt(sum((BWkbs-Wkbs[indk])**2)/B)
sk = sk*np.sqrt(1+1/B)
return(ks, Wks, Wkbs, sk)